Insite - In-Situ Pipeline REST API (2.1.0)
Download OpenAPI specification:Download
This research was supported by the EBRAINS research infrastructure, funded from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 945539 (Human Brain Project SGA3). This project/research has received funding from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 945539 (Human Brain Project SGA3) and Specific Grant Agreement No. 785907 (Human Brain Project SGA2).
Insite provides a middleware that enables users to acquire data from neural simulators via the in-transit paradigm. In-transit approaches allow users to access data from a running simulation while the simulation is still going on. In the traditional approach data from simulations is written to disk first, and can only be accessed after the simulation has finished. However, this has two main constraints:
- Data can only be further processed after the whole simulation has finished.
- Disk speed can be a bottleneck when simulating, as data has to be written out.
- Data must be completely stored on the machine, leading to large files.
Using Insite allows users to develop data consumer, such as visualizations and analysis tools that allow early insight into the data without storing data with virtually zero dependencies.
Insite was specifically designed to be ease-to-integrate and easy-to-use to allow a wide range of users to take advantage of in-transit approaches in the context of brain simulatiion. Insite uses off-the-shelf dataformats and protocols to make integration as easy as possible. Data can be queried via an HTTP REST API from Insite's access node, which represents a single point of contact for the user.
Insite consists of two main layers. The first part is the Insite access node, an HTTP server that serves as a centralized endpoints.
The access node aggregates data from multiple simulators (NEST, Arbor, TVB) and from multiple simulation instances, respectively. This means, that even distributed simulations, e.g. via MPI, can be access as if it is only one simulation instances. Data is aggregated at the access node and can then be filtered and accessed by the user. Thus, allowing convienent access to multi-node multiscale simulations.
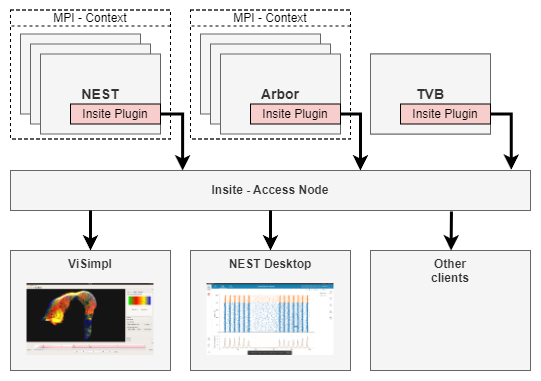
This architecture enables users to easily create in-situ visualizations of neural simulations without the knowledge of the simulation's underlying architecture or any further knowledge about the computational distribution of the simulation. Users can query the HTTP endpoint for different datapoints, such as: spike data, multimeter data, the topology of the neural network and so on (see full API below). The returned data in encoded in JSON, thus allowing processing in a plethora of different languages and systems without Insite specific dependencies.
The access-node provides the full API to the Insite pipeline which is documented below.
The second part are Insite's simulator plugings. The simulator plugin is embedded into the respective simulations and is responsible for collecting simulation data and making it available to the access node. The simulators plugins are designed to be as non-invasive as possible to make integration into new and existing simulations easy. Simulator plugins are available for NEST, Arbor and The Virtual Brain.
Additional information can be found on the following poster:
To be able to use the Docker images please make sure that the docker engine and docker compose are installed.
(Or docker compatible containerization engines)
There are two different approaches for using Insite with docker:
Run the example
The docker-compose below provides an example with all three simulators.
While in this case each simulator runs their own simulation, all data can be queried via the single access-node nonetheless.
Just save the docker-compose as some-name.yaml and then start Insite with docker-compose -f some-name.yaml up.
Docker will download all four containers and start the simulations.
You can query the data from the examples from the access node via localhost:52056 via your favourite tool, e.g.: curl -L localhost:52056/version/ to retrieve the version.
version: "3"
services:
insite-access-node:
image:
docker-registry.ebrains.eu/insite/access-node
ports:
- "52056:52056" # You can change the local part of the port here
environment:
INSITE_NEST_BASE_URL: "insite-nest-example"
INSITE_ARBOR_BASE_URL: "insite-arbor-example"
insite-nest-example:
image:
docker-registry.ebrains.eu/insite/insite-nest-example
depends_on:
- insite-access-node
insite-tvb-example:
image:
docker-registry.ebrains.eu/insite/insite-tvb-example
environment:
INSITE_ACCESS_NODE_URL: "insite-access-node"
depends_on:
- insite-access-node
insite-arbor-example:
image:
docker-registry.ebrains.eu/insite/insite-arbor-example
depends_on:
- insite-access-node
Using Pre-built Images
We provide pre-built images for NEST, Arbor, TVB and the Insite access node. These images contain the respective simulator binaries and the Insite simulator module pre-installed:
| Container | Link |
|---|---|
| Arbor | Link |
| NEST | Link |
| TVB | Link |
| Access Node | Link |
You can use these images to deploy your own Insite-enabled simulations. For further information on how to that you can refer to our examples.
Build Docker Images
To build the docker containers yourself you can use the provided Dockerfile in the repository.
Dockerfiles are located in the docker with the following structure:
docker
├── Dockerfile_AccessNode
├── examples
│ ├── Dockerfile_ArborRingExample
│ ├── Dockerfile_NestExample
│ └── Dockerfile_TVBExample
└── simulators
├── Dockerfile_Arbor
├── Dockerfile_NEST
└── Dockerfile_TVB
Dockerfile_AccessNode builds Insite's access node.
simulators/Dockerfile_* builds the respective simulators with Insite simulator modules integrated.
examples/Dockerfile_* builds the respective examples. They are based on the simulator docker images but add example simulations.
The Dockerfiles refer to relative locations starting at the repository root. Therefore, to build containers please build from the root and refer to the Dockerfiles, e.g.:
docker build . -f docker/simulators/Dockerfile_Arbor
- Clone this repository (
git clone https://github.com/VRGroupRWTH/insite.git).
Building the Access Node
The access node is written in C++ and uses the CMake build system. Dependencies are downloaded via CPM during configuration.
cd access-node
mkdir build && cd build
cmake ..
make -j insite-access-node
Access node dependencies:
- https://github.com/jbeder/yaml-cpp
- https://github.com/libcpr/cpr
- https://github.com/zaphoyd/websocketpp
- https://github.com/gabime/spdlog
- https://github.com/google/flatbuffers
- https://github.com/marzer/tomlplusplus
- https://github.com/OlivierLDff/asio.cmake
- https://github.com/TartanLlama/optional
- https://github.com/Tencent/rapidjson
- https://github.com/CrowCpp/Crow
Building the Arbor Module
The arbor module is written in C++ and uses the CMake build system. Dependencies are downloaded via CPM during configuration.
cd arbor-module
mkdir build && cd build
cmake ..
make -j insite-arbor
make install
Arbor module dependencies:
- https://github.com/arbor-sim/arbor
- https://github.com/gabime/spdlog
- https://github.com/marzer/tomlplusplus
- https://github.com/TartanLlama/optional
- https://github.com/Tencent/rapidjson
- https://github.com/CrowCpp/Crow
Building the NEST Module
The nest module is written in C++ and uses the CMake build system. Dependencies are downloaded via CPM during configuration. First build nest according to the the nest documentation, then build against nest.
cd nest-module
mkdir build && cd build
cmake .. -Dwith-nest=NEST_INSTALL_PATH/bin/nest-config
make -j install
NEST module dependencies:
- https://github.com/nest/nest-simulator
- https://github.com/zaphoyd/websocketpp
- https://github.com/gabime/spdlog
- https://github.com/google/flatbuffers
- https://github.com/TartanLlama/optional
- https://github.com/Tencent/rapidjson
- https://github.com/CrowCpp/Crow
Building the TVB module
The TVB module is written in Python and involves no compilation process. Dependencies can be installed via pip:
cd tvb-module
pip install -r requirements.txt
Tvb module dependencies:
The following sections assumes that the Insite nest module was successfully compiled and installed and is available.
Either through using the NEST insite container described above or by manual compilation.
Insite can be integrated into NEST simulation via NEST's Python API.
A NEST simulation can be in-transit enabled by adding Insite recording backends into the simulation after activating Insite with:
nest.Install("insitemodule")
Afterwards, a new spike recorder or multimeter can be created with:
# Create spike recorder
insite_spike_recorder = nest.Create("spike_recorder")
#Create multimeter
insite_multimeter = nest.Create("multimeter")
To enable recording to Insite the spikerecorder and multimeter have to be set to the correct recording backend with:
nest.SetStatus(YOUR_DEVICE, [{"record_to":"insite"}])
respectively.
The recording devices act as normal NEST nodes and can be connected to other nodes, e.g., neurons:
nest.Connect(nodes_ex, insite_spike_recorder, syn_spec="excitatory")
All neurons connected to Insite devices are available via the Insite pipeline.
Endpoints for spikerecorders and multimeters provide information about Insite-enabled nodes in the network while the spike and multimeter endpoints return data recorded by the devices.
The following sections assumes that the Insite nest module was successfully compiled and installed and is available.
Either through using the Arbor insite container described above or by manual compilation.
Insite can be embedded into Arbor simulations via the Insite arbor C++ module.
Therefore, arbor simulations using Insite must link against insite-arbor in their CMakeLists. E.g.:
target_link_libraries(YOUR_SIMULATION PUBLIC insite-arbor)
Afterwards, the simulation can be made Insite-enabled by embedding the Insite pipeline backend into your simulation file.
First include the pipeline backend:
#include <insite/arbor/pipeline_backend.h>
Next, add an Insite HTTP backend in your main code before running the simulation:
insite::HttpBackend insite_backend(context, recipe, sim, arb::all_probes,
arb::regular_schedule(1.0));
The signature is as follows:
HttpBackend(const arb::context& ctx,
const arb::recipe& rcp,
arb::simulation& sim,
arb::cell_member_predicate cell_pred,
arb::schedule sched);
The cell_member_predicate can be used to filter the probes that should be made available via Insite.
In the example arb::all_probes enables all present probes to be accessed via Insite.
The sched describes the sampling rate of the Insite-enabled probes.
The Backend provides information about the cells present in the simulation. Additionally, the spikes of all neurons are recorded by default. Available probes and their data can be queried via the API.
Please refer to the API documentation below for a complete overview.
The following sections assumes that the Insite TVB module is available.
Either through using the Arbor insite container described above or by manual installation.
First import the Python TVB modules:
from insite_websocketmanager import InsiteWebSocketManager, InsiteOpcode
from insite_monitor_wrapper import InsiteMonitorWrapper
afterwards initialize the InsiteWebSocketManager:
insite_ws_manager = WebSocketManager(INSITE_ACCESS_NODE_URL,9011)
the access node url.
To record data to Insite regular monitors have to be wrapped via the InsiteMonitorWrapper:
mon_raw = monitors.Raw()
mon_spat = monitors.SpatialAverage()
insite_spatial = MonitorWrapper(mon_spat,insite_ws_manager)
insite_raw = MonitorWrapper(mon_raw,insite_ws_manager)
Monitors can be configured as usual before calling the MonitorWrapper. All wrapped monitors that are registered as monitors at the simulation will be will be available via Insite pipeline.
what_to_watch = [insite_raw, insite_spatial]
sim = simulator.Simulator(model = oscilator, connectivity = white_matter,
coupling = white_matter_coupling,
integrator = heunint, monitors = what_to_watch)
Before starting the simulation connect() and the send_start_sim() function must be called.
send_sim_info(sim) must be called before configure is called on the simulation.
After the simulation send_end_sim() must be called.
Finally, the connection can be close via the close() call.
The following code shows the complete boilerplate code for wrapping a TVB simulation:
insite_ws_manager = WebSocketManager(address,9011)
insite_ws_manager.connect()
mon_raw = monitors.Raw()
mon_spat = monitors.SpatialAverage()
insite_spatial = MonitorWrapper(mon_spat,insite_ws_manager)
insite_raw = MonitorWrapper(mon_raw,insite_ws_manager)
length = 500
what_to_watch = [insite_spatial]
[simulation settings...]
insite_ws_manager.send_start_sim()
sim = simulator.Simulator(model = oscilator, connectivity = white_matter,
coupling = white_matter_coupling,
integrator = heunint, monitors = what_to_watch)
insite_ws_manager.send_sim_info(sim)
sim.configure()
for data in sim(simulation_length=length):
pass
insite_ws_manager.send_end_sim()
insite_ws_manager.close()
The following settings can be configured on the access node:
| Description | config key | env var |
|---|---|---|
| #NEST Nodes | numberOfNodes | INSITE_NEST_NUM_NODES |
| URL of NEST Simulator | baseUrl | INSITE_NEST_BASE_URL |
| #Arbor Nodes | numberOfNodes | INSITE_ARBOR_NUM_NODES |
| URL of Arbor Simulator | baseUrl | INSITE_ARBOR_BASE_URL |
| Access Node API Port | port | INSITE_ACCESS_NODE_PORT |
| Access Node Websocket Port (TVB Port) | websocketPort | INSITE_ACCESS_NODE_WSPORT |
Values can be provided by settings the corresponding environment variable or via TOML or YAML in a config.{toml,yaml} file via the same directory as the binary.
Toml example:
["nest"]
numberOfNodes = 2
baseUrl = "insite-nest-example"
["arbor"]
numberOfNodes = 1
baseUrl = "insite-arbor-example"
["accessNode"]
port = 52056
websocketPort = 9011
Yaml example:
nest:
numberOfNodes: 2
baseUrl: "insite-nest-example"
arbor:
numberOfNodes: 1
baseUrl: "insite-arbor-example"
accessNode:
port: 52056
websocketPort: 9011
Release 2.1 of Insite has support for the following simulators:
- NEST
- (new) Arbor
- (new) TVB and incorporates the following new components:
- C++ Arbor simulator module
- Python TVB simulator module
- TVB integration tests
- Arbor integration tests
- Docker support:
- Arbor container with Insite example
- TVB container with Insite example
- docker-compose file incorporating all three simulators and Access Node enhanced components:
- C++ NEST simulator module
- C++ Access Node
- Docker support:
- Access Node container
- NEST container with Insite example
- Access Node container as well as:
- Miscellaneous bug fixes
- Updated Documentation
- Integration into Ebrains CI/CD
Release 2.0 of Insite has support for the following simulators:
- NEST and incorporates the following enhancements:
- Complete rewrite of the Access Node into C++
- General performance improvements
- Parallalization of requests to simulation nodes
NOTE: 2.0 release scraped in benefit of merge with 2.1
Release 1.1 of Insite has support for the following simulators:
- NEST and incorporates the following enhancements:
- C++ NEST simulator module updated for NEST 3.2
- Major performance improvements for Python Access Node
- Improved API:
- Adds fields to indicate that requests contain the last data point
- Adds fields to indicate that simulation is running or has finished
- Compatibility to NEST Server
- Improved integration tests
- Miscellaneous bug fixes
Release 1.0 of Insite has support for the following simulators:
- NEST and incorporates the following components:
- C++ NEST simulator module
- Python Access Node
- Docker containers:
- Access Node
- NEST Simulator with Insite module installed
Monitor Data
Returns recorded data from the monitors in the TVB instance that are connected to Insite and available to fetch data from.
query Parameters
| gId | integer <uint64> One or more gIds that should be filtered for. |
Responses
Response samples
- 200
[- {
- "uid": "467bd4ff-3625-4440-9b3a-e7395cab5cc7",
- "timesteps": [
- {
- "time": 499.0234375,
- "V": [
- -0.19128321122285258
], - "W": [
- -0.08729334745723864
], - "V + W": [
- -0.2785765586800911
]
}, - {
- "time": 500,
- "V": [
- -0.19128247282170338
], - "W": [
- -0.0872909920743438
], - "V + W": [
- -0.2785734648960471
]
}
]
}
]Simulation Properties
Returns the simulation properties of the TVB instance connected to the the Insite instance. Returned values can be inspected further with the /simulation_info/
Responses
Response samples
- 200
{- "Type": "Simulator",
- "title": "Simulator gid: e0f27575-bc8c-4444-a298-0bd7a272ac43",
- "connectivity": "Connectivity gid: de59731a-f13f-4103-a2d5-9182eb4ba213",
- "conduction_speed": "3.0",
- "coupling": "Linear gid: 520ba213-dee2-41c8-972d-95ffc71f6d38",
- "surface": "None",
- "stimulus": "None",
- "model": "Generic2dOscillator gid: de979767-c3fb-4072-a8a0-5ff096b5ef21",
- "integrator": "HeunDeterministic gid: 71890b28-7f4e-4cfa-a45c-ecb7ba92ef1a",
- "initial_conditions": "None",
- "monitors": "[<monitor_wrapper.MonitorWrapper object at 0x7fc7c94b5210>]",
- "simulation_length": "1000.0",
- "gid": "UUID('e0f27575-bc8c-4444-a298-0bd7a272ac43')"
}Subproperties of Simulation Properties
Returns the simulation subproperties of the TVB instance connected to the the Insite instance. Returned values can be inspected further with the /simulation_info/
path Parameters
| property required | any Name of a property returned by /simulation_info or /simulation_info/other_prorperty. |
Responses
Response samples
- 200
{- "Number of regions": 76,
- "Number of connections": 1560,
- "Undirected": false,
- "areas": " [min, median, max] = [0, 2580.89, 10338.2] dtype = float64 shape = (76,)",
- "weights": " [min, median, max] = [0, 0, 3] dtype = float64 shape = (76, 76)",
- "weights-non-zero": " [min, median, max] = [0.00462632, 2, 3] dtype = float64 shape = (1560,)",
- "tract_lengths": " [min, median, max] = [0, 71.6635, 153.486] dtype = float64 shape = (76, 76)",
- "tract_lengths-non-zero": " [min, median, max] = [4.93328, 74.0646, 153.486] dtype = float64 shape = (5402,)",
- "tract_lengths (connections)": " [min, median, max] = [0, 55.8574, 138.454] dtype = float64 shape = (1560,)"
}Simulation Probes
Returns all probes that are present in the simulation and are connected to the Insite instance.
query Parameters
| gId | integer <uint64> One or more gIds that should be filtered for. |
| lId | integer <uint64> One or more lIds that should be filtered for. |
| pId | integer <uint64> One or more pIds that should be filtered for. |
| uId | integer <uint64> One or more uIds that should be filtered for. |
| hash | integer <uint64> One or more probe hashes that should be filtered for. |
Responses
Response samples
- 200
{- "probes": [
- {
- "cell_gid": 2,
- "cell_lid": 1,
- "source_index": 0,
- "probe_kind": "cable_probe_axial_current",
- "hash": 4294967298,
- "probe_global_index": 11,
- "location": [
- 0,
- 0.5
]
}, - {
- "cell_gid": 2,
- "cell_lid": 0,
- "source_index": 2,
- "probe_kind": "cable_probe_membrane_voltage",
- "hash": 562949953421314,
- "probe_global_index": 10,
- "location": [
- 0,
- 0.3
]
}
]
}Probe Data
Returns data from the probes that are present in the simulation and are connected to the Insite instance.
query Parameters
| gId | integer <uint64> One or more gIds that should be filtered for. |
| lId | integer <uint64> One or more lIds that should be filtered for. |
| pId | integer <uint64> One or more pIds that should be filtered for. |
| uId | integer <uint64> One or more uIds that should be filtered for. |
| hash | integer <uint64> One or more probe hashes that should be filtered for. |
| fromTime | number <double> The start time in milliseconds (including) to be queried. |
Responses
Response samples
- 200
{- "probeData": [
- {
- "gid": 2,
- "lid": 1,
- "pid": 0,
- "size": 1000,
- "time": [
- 0,
- 0.9750000000000004,
- 1.999999999999997,
- 2.9999999999999933,
- 3.99999999999999
], - "data": [
- -1.1013412404281553e-13,
- 0.014220165459628968,
- 0.012908425336631052,
- 0.012920058448978668,
- 0.013377748739968996
]
}, - {
- "gid": 2,
- "lid": 0,
- "pid": 2,
- "size": 1000,
- "time": [
- 0,
- 0.9750000000000004,
- 1.999999999999997,
- 2.9999999999999933,
- 3.99999999999999
], - "data": [
- -65,
- -67.31833822951484,
- -68.46917744645607,
- -68.89870365000968,
- -69.03936729932849,
- -69.06695156902677
]
}
]
}Spikes
Returns spikes that occured in the simulation.
query Parameters
| fromTime | number <double> The start time in milliseconds (including) to be queried. |
| toTime | number <double> The end time in milliseconds (excluding) to be queried. |
| gId | integer <uint64> One or more gIds that should be filtered for. |
| skip | integer <uint64> The offset into the result. |
| top | integer <uint64> The maximum number of entries to be returned. |
Responses
Response samples
- 200
{- "simulationTimes": [
- 0.8,
- 0.9
], - "gIds": [
- 1,
- 2
]
}Cells
Returns the cells and their properties that are present in the connected Arbor simulation.
Responses
Response samples
- 200
{- "cells": [
- {
- "gid": 0,
- "cell_kind": "cable",
- "cell_description": {
- "morphology": {
- "branches": [
- [
- "(morphology\n ((4294967295 ((segment 0 (point 0 0 -6.30785 6.30785) (point 0 0 6.30785 6.30785) 1)))\n (0 ((segment 1 (point 0 0 6.30785 6.30785) (point 0 0 16.3079 0.5) 3)"
]
]
}, - "label": {
- "regions": [
- {
- "name": "dend",
- "region": "(tag 3)"
}, - {
- "name": "soma",
- "region": "(tag 1)"
}
], - "locset": [ ]
}, - "decor": {
- "painting": [
- {
- "region": "(region \"soma\")",
- "paintable": {
- "type": "density",
- "value": "hh"
}
}, - {
- "region": "(region \"dend\")",
- "paintable": {
- "type": "density",
- "value": "pas"
}
}
], - "placement": [
- {
- "region": "(location 0 0)",
- "placeable": {
- "label": "detector",
- "type": "threshold_detector"
}
}, - {
- "region": "(location 0 0.5)",
- "placeable": {
- "label": "primary_syn",
- "type": "synapse"
}, - "value": "expsyn"
}
]
}
}
}
]
}Response samples
- 200
{- "CellInfos": [
- {
- "gid": 0,
- "cell_kind": "cable",
- "num_branches": 19,
- "num_segments": 231
}, - {
- "gid": 1,
- "cell_kind": "cable",
- "num_branches": 13,
- "num_segments": 174
}, - {
- "gid": 2,
- "cell_kind": "cable",
- "num_branches": 19,
- "num_segments": 216
}
]
}Response samples
- 200
[- {
- "T_max": 1152921504606846.8,
- "T_min": -1152921504606846.8,
- "adaptive_spike_buffers": true,
- "adaptive_target_buffers": true,
- "buffer_size_secondary_events": 0,
- "buffer_size_spike_data": 42,
- "buffer_size_target_data": 125,
- "data_path": "",
- "data_prefix": "",
- "dict_miss_is_error": true,
- "grng_seed": 0,
- "growth_factor_buffer_spike_data": 1.5,
- "growth_factor_buffer_target_data": 1.5,
- "keep_source_table": true,
- "local_num_threads": 1,
- "local_spike_counter": 745,
- "max_buffer_size_spike_data": 8388608,
- "max_buffer_size_target_data": 16777216,
- "max_delay": 1.5,
- "max_num_syn_models": 512,
- "min_delay": 1.5,
- "ms_per_tic": 0.001,
- "network_size": 128,
- "num_connections": 1750,
- "num_processes": 1,
- "off_grid_spiking": false,
- "overwrite_files": true,
- "print_time": false,
- "recording_backends": {
- "ascii": { },
- "insite": { },
- "memory": { },
- "screen": { }
}, - "resolution": 0.1,
- "rng_seeds": [
- 1
], - "sort_connections_by_source": true,
- "structural_plasticity_synapses": { },
- "structural_plasticity_update_interval": 10000,
- "tics_per_ms": 1000,
- "tics_per_step": 100,
- "time": 100,
- "time_collocate": 0,
- "time_communicate": 0,
- "to_do": 0,
- "total_num_virtual_procs": 1,
- "use_wfr": true,
- "wfr_comm_interval": 1,
- "wfr_interpolation_order": 3,
- "wfr_max_iterations": 15,
- "wfr_tol": 0.0001
}
]Multimeters
Retrieves a list with information about all available multimeters and their properties. Additionally, the ids of the nodes that are connected to the multimeter are returned.
Responses
Response samples
- 200
[- {
- "multimeterId": 10,
- "attributes": [
- "V_m"
], - "nodeIds": [
- 1,
- 3,
- 5
]
}
]Multimeter Properties
Retrieves information about a specific multimeter that and its properties.
path Parameters
| multimeterId required | integer <uint64> The identifier of the multimeter. |
Responses
Response samples
- 200
{- "multimeterId": 10,
- "attributes": [
- "V_m"
], - "nodeIds": [
- 1,
- 3,
- 5
]
}Mutlimeter Measurements By MultimeterId
path Parameters
| multimeterId required | integer <uint64> The multimeter to query |
| attributeName required | string The attribute to query (e.g., 'V_m' for the membrane potential) |
query Parameters
| fromTime | number <double> The start time (including) to be queried. |
| toTime | number <double> The end time (excluding) to be queried. |
| nodeIds | Array of integers <uint64> [ items <uint64 > ] A list of node IDs queried for attribute data. |
| skip | integer <uint64> The offset into the result. |
| top | integer <uint64> The maximum number of entries to be returned. |
Responses
Response samples
- 200
{- "simulationTimes": [
- 0.1,
- 0.2
], - "nodeIds": [
- 1,
- 2,
- 3
], - "values": [
- 0.123,
- 0.123,
- 0.123,
- 0.123,
- 0.123,
- 0.123
]
}Node Collections
Retrieves an list of all node collections. Returned are the node collection IDs, which nodes are part of the collection and which model is used by the collection.
Responses
Response samples
- 200
[- {
- "nodeCollectionId": 0,
- "nodes": {
- "firstId": 1,
- "lastId": 100,
- "count": 100
}, - "model": {
- "name": "iaf_psc_delta",
- "status": {
- "C_m": "1,",
- "Ca": "0,",
- "E_L": "0,",
- "I_e": "0,",
- "V_m": "0,",
- "V_min": "-1.7976931348623157e+308,",
- "V_reset": "0,",
- "V_th": "20,"
}
}
}
]Nodes In Node Collection
Retrieves the list of all nodes within the specified node collection.
path Parameters
| nodeCollectionId required | integer <uint64> The identifier of the node collection |
Responses
Response samples
- 200
[- {
- "nodeId": 1,
- "nodeCollectionId": 0,
- "simulationNodeId": 0,
- "position": [
- 0.1,
- 0.2
], - "model": "iaf_psc_delta",
- "status": {
- "C_m": 1,
- "Ca": 0,
- "E_L": 0,
- "I_e": 0,
- "V_m": 0,
- "V_min": -1.7976931348623157e+308,
- "V_reset": 0,
- "V_th": 20,
- "archiver_length": 0,
- "beta_Ca": 0.001
}
}
]Spikes By Node Collection ID
Retrieves the spikes for the given simulation steps (optional) and node collection. This request merges the spikes recorded by all spike detectors and removes duplicates.
path Parameters
| nodeCollectionId required | integer <uint64> The identifier of the node collection. |
query Parameters
| fromTime | number <double> The start time (including) to be queried. |
| toTime | number <double> The end time (excluding) to be queried. |
| skip | integer <uint64> The offset into the result. |
| top | integer <uint64> The maximum numbers of entries to be returned. |
Responses
Response samples
- 200
{- "simulationTimes": [
- 0.8,
- 0.9
], - "nodeIds": [
- 1,
- 2
], - "lastFrame": false
}Response samples
- 200
[- {
- "nodeId": 1,
- "nodeCollectionId": 0,
- "simulationNodeId": 0,
- "position": [
- 0.1,
- 0.2
], - "model": "iaf_psc_delta",
- "status": {
- "C_m": 1,
- "Ca": 0,
- "E_L": 0,
- "I_e": 0,
- "V_m": 0,
- "V_min": -1.7976931348623157e+308,
- "V_reset": 0,
- "V_th": 20,
- "archiver_length": 0,
- "beta_Ca": 0.001
}
}
]Node Properties By Node ID
Retrieves the properties of the specified node.
path Parameters
| nodeId required | integer <uint64> The ID of the queried node. |
Responses
Response samples
- 200
{- "nodeId": 1,
- "nodeCollectionId": 0,
- "simulationNodeId": 0,
- "position": [
- 0.1,
- 0.2
], - "model": "iaf_psc_delta",
- "status": {
- "C_m": 1,
- "Ca": 0,
- "E_L": 0,
- "I_e": 0,
- "V_m": 0,
- "V_min": -1.7976931348623157e+308,
- "V_reset": 0,
- "V_th": 20,
- "archiver_length": 0,
- "beta_Ca": 0.001
}
}Spikes By Spike Recorder Id
Retrieves the spikes for the given time range (optional) and node IDs (optional) from one spike recorder. If no time range or node list is specified, it will return the spikes for whole time or all nodes respectively.
path Parameters
| spikerecorderId required | integer <uint64> The ID of the spike detector to query. |
query Parameters
| fromTime | number <double> The start time in milliseconds (including) to be queried. |
| toTime | number <double> The end time in milliseconds (excluding) to be queried. |
| nodeIds | Array of integers <uint64> [ items <uint64 > ] A list of node IDs queried for spike data. |
| skip | integer <uint64> The offset into the result. |
| top | integer <uint64> The maximum number of entries to be returned. |
Responses
Response samples
- 200
{- "simulationTimes": [
- 0.8,
- 0.9
], - "nodeIds": [
- 1,
- 2
], - "lastFrame": false
}Spikes By Spikedetector Id Deprecated
Retrieves the spikes for the given time range (optional) and node IDs (optional) from one spike detector. If no time range or node list is specified, it will return the spikes for whole time or all nodes respectively.
path Parameters
| spikedetectorId required | integer <uint64> The ID of the spike detector to query. |
query Parameters
| fromTime | number <double> The start time in milliseconds (including) to be queried. |
| toTime | number <double> The end time in milliseconds (excluding) to be queried. |
| nodeIds | Array of integers <uint64> [ items <uint64 > ] A list of node IDs queried for spike data. |
| skip | integer <uint64> The offset into the result. |
| top | integer <uint64> The maximum number of entries to be returned. |
Responses
Response samples
- 200
{- "simulationTimes": [
- 0.8,
- 0.9
], - "nodeIds": [
- 1,
- 2
], - "lastFrame": false
}Spikes
Retrieves the spikes for the given time range (optional) and node IDs (optional). If no time range or node list is specified, it will return the spikes for whole time or all nodes respectively. This request merges the spikes recorded by all spike detectors and removes duplicates.
query Parameters
| fromTime | number <double> The start time in milliseconds (including) to be queried. |
| toTime | number <double> The end time in milliseconds (excluding) to be queried. |
| nodeIds | Array of integers <uint64> [ items <uint64 > ] A list of node IDs queried for spike data. |
| skip | integer <uint64> The offset into the result. |
| top | integer <uint64> The maximum number of entries to be returned. |
Responses
Response samples
- 200
{- "simulationTimes": [
- 0.8,
- 0.9
], - "nodeIds": [
- 1,
- 2
], - "lastFrame": false
}